Supplementary files and notes of the manuscript
Pramanik et al., 2020: Proteomic Atomics Reveals a Distinctive Uracil-5-Methyltransferase
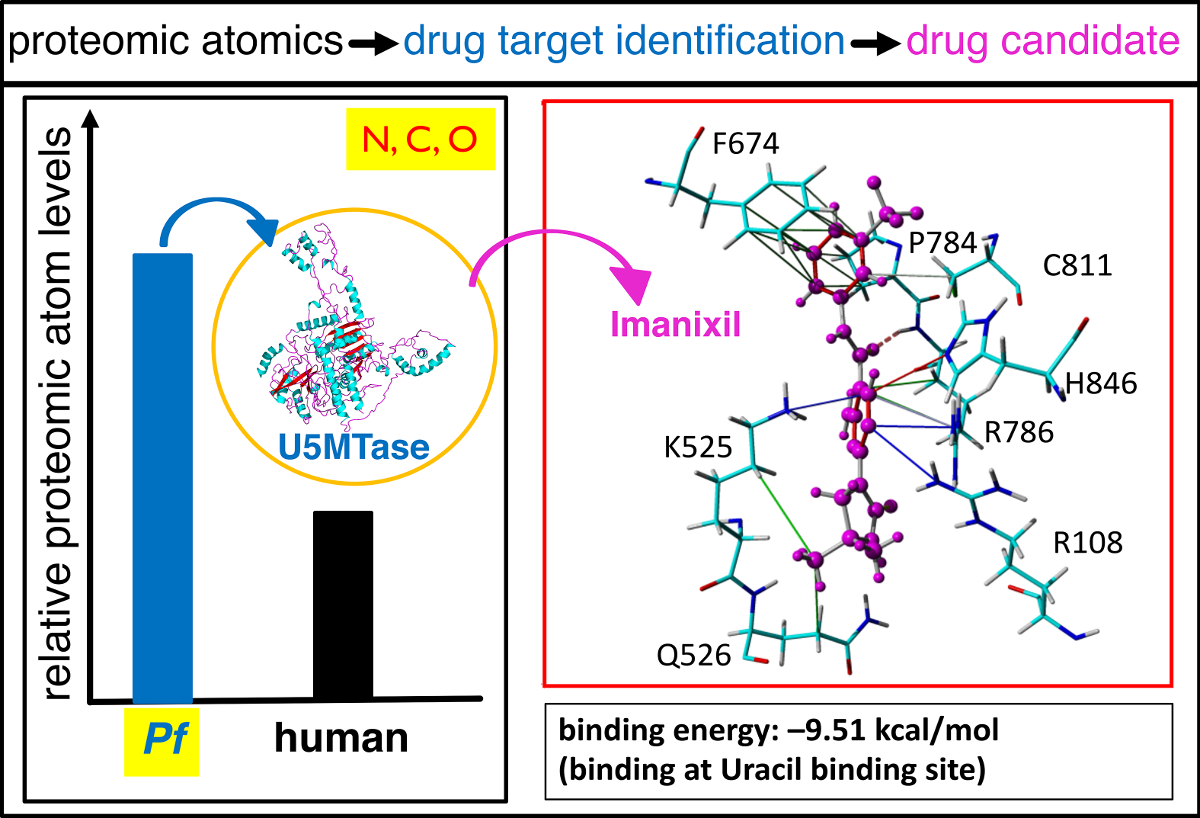
Figure: Proteome-based atom (C, H, N, O, S) distributions across all species of the phylogenetic tree revealed U5MTase of Plasmodium falciparum as a distinguished possible therapeutic target which in turn was used for in silico structure-based drug design strategies (i.e. 3D protein structure modeling, virtual chemical library screening, and molecular docking) to identify imanixil as potential inhibitory drug molecule that might serve as possible remedy for the treatment of malaria.
PDB Files
Below you find the 3D protein structure pdb files mentioned in the manuscript:
- U5MTAse+SAM: U5MTAse+SAM
- U5MTase+Uracil: U5MTase+Uracil.pdb
- U5MTAse+Zn2+: U5MTAse+Zn2+.pdb
- U5MTase+Zn_SAM + Imanixil: U5MTase+Zn_SAM + Imanixil.pdb
- U5MTase+Zn_SAM + Tannin: U5MTase+Zn_SAM + Tannin.pdb
- U5MTase+Zn_SAM + Encalaret: U5MTase+Zn_SAM + Encalaret.pdb
>>>> Uracil-5-methyltransferase revisited – a suitable target for malaria?
Software tools
For the CARd software tool, please address your inquiries directly to Prof. Rajasekaran Ekambaram: <ersekaran@gmail.com>
References
Proteomic Atomics Reveals a Distinctive Uracil-5-Methyltransferase.
Pramanik S, Thaker M, Perumal AG, Ekambaram R, Poondla N, Schmidt M, Kim PS, Kutzner A, Heese K#.
Molecular Informatics xxx: y-z (2020). (*2.3) (SCI) (in press).
Oxygen distribution in proteins defines functional significance of the genome and proteome of the malaria parasite Plasmodium falciparum 3D7.
Palanisamy B, Heese K.
FEMS Microbiol Lett. 2014; 351(1): 59-63.
Thymine distribution in genes provides novel insight into the functional significance of the proteome of the malaria parasite Plasmodium falciparum 3D7.
Palanisamy B, Ekambaram R, Heese K.
Bioinformatics. 2014; 30(5): 597-600.